from read_pdz import extract_spectrumExtracting spectral data from pdz files
This quote by internet pioneer Marleen Stikker is a fundamental truth that applies to software and technology as well as to data. In my lab there are many examples of data file formats that we can not easily read, share and use. This needs to change.
Bruker .pdz files are binary files that contain both XRF spectral data (i.e. photon counts) and metadata. Let’s ignore the gory details for now and show how to extract the spectral data and make a plot of the spectrum or spectra that are hidden in the file. To do so import the extract_spectra() function from the read_pdz package. The function requires a valid file path to a pdz file. It returns a pandas DataFrame that can be inspected in the notebook and is automatically saved as a .csv file in the same folder as the pdz file.
pdz_file = '/home/frank/Work/DATA/pdz-vgm-example-with-exports/demo.pdz'
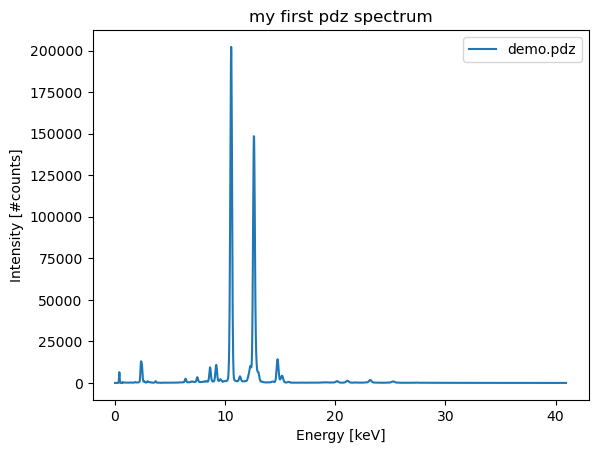
spectrum_df = extract_spectrum(pdz_file)Saving spectral data to: /home/frank/Work/DATA/pdz-vgm-example-with-exports/demo.pdz.csvThe spectral data in the DataFrame can now be processed further. If you quickly want to inspect the spectra you can use the .plot() method.
ax = spectrum_df.plot()
ax.set_title('my first pdz spectrum')
ax.set_xlabel('Energy [keV]')
ax.set_ylabel('Intensity [#counts]');